Using RNA-Seq to Explore TranscriptionAl Changes During Axon Regeneration
This project grew out of the PNS vs CNS Regeneration project. Instead of using subtractive hybridization to explore the transcriptomic differences between two cell types, we are now using the newer technology of RNA-Seq. We first applied this method as an extension of the subtractive hybridization experiment to look at the transcriptional and isoform differences between Dorsal Root Ganglion (DRG) neurons and Cerebellar Granule Neurons (CGN) in culture (Lerch JK, et al. 2012). Next, we performed RNA-Seq on RNA that had been isolated by laser capture microdissection (LCM) from DRGs that had either undergone a sciatic nerve crush or a sham surgery in order to observe what transcriptional changes occurred in DRGs as they responded to injury and regenerated their axons. Utilization of these cutting edge techniques allows us to answer questions about development and regeneration on a scale never before possible.
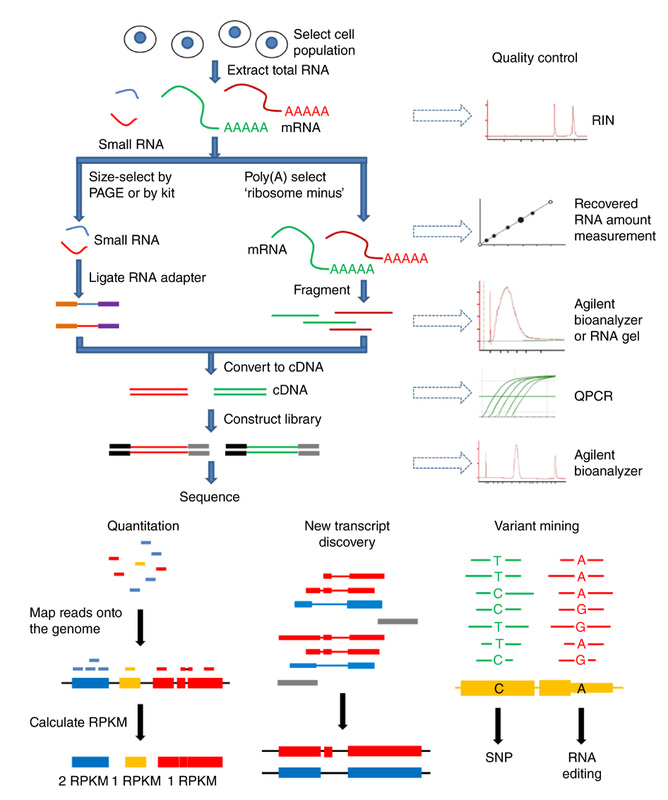
Figure from Zeng, W & Mortazavi, A (2012) Nature Immunology 13, 802-807.
References:
Lerch JK, Kuo F, Motti D, Morris R, Bixby JL, Lemmon VP. Isoform diversity and regulation in peripheral and central neurons revealed through RNA-Seq. PLoS One. 2012;7:e30417. doi: 10.1371/journal.pone.0030417.
References:
Lerch JK, Kuo F, Motti D, Morris R, Bixby JL, Lemmon VP. Isoform diversity and regulation in peripheral and central neurons revealed through RNA-Seq. PLoS One. 2012;7:e30417. doi: 10.1371/journal.pone.0030417.
| plos_one_2012_lerch.pdf |